Medicine is an inherently multimodal discipline. When providing care, clinicians routinely interpret data from a wide range of modalities including medical images, clinical notes, lab tests, electronic health records, genomics, and more. Over the last decade or so, AI systems have achieved expert-level performance on specific tasks within specific modalities — some AI systems processing CT scans, while others analyzing high magnification pathology slides, and still others hunting for rare genetic variations. The inputs to these systems tend to be complex data such as images, and they typically provide structured outputs, whether in the form of discrete grades or dense image segmentation masks. In parallel, the capacities and capabilities of large language models (LLMs) have become so advanced that they have demonstrated comprehension and expertise in medical knowledge by both interpreting and responding in plain language. But how do we bring these capabilities together to build medical AI systems that can leverage information from all these sources?
In today’s blog post, we outline a spectrum of approaches to bringing multimodal capabilities to LLMs and share some exciting results on the tractability of building multimodal medical LLMs, as described in three recent research papers. The papers, in turn, outline how to introduce de novo modalities to an LLM, how to graft a state-of-the-art medical imaging foundation model onto a conversational LLM, and first steps towards building a truly generalist multimodal medical AI system. If successfully matured, multimodal medical LLMs might serve as the basis of new assistive technologies spanning professional medicine, medical research, and consumer applications. As with our prior work, we emphasize the need for careful evaluation of these technologies in collaboration with the medical community and healthcare ecosystem.
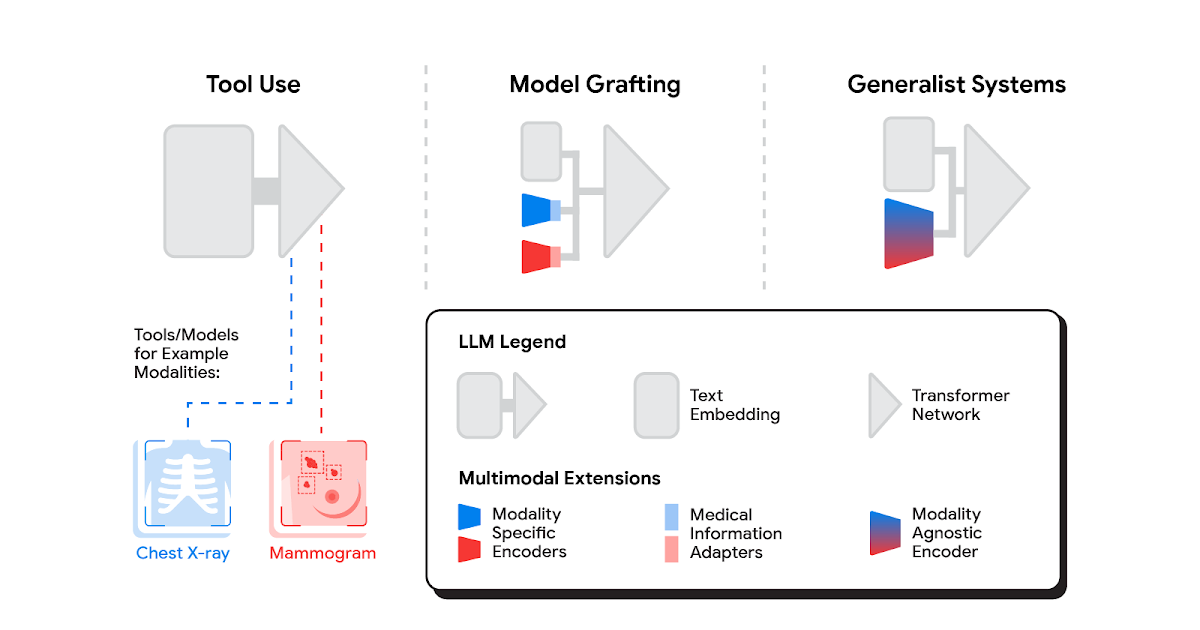
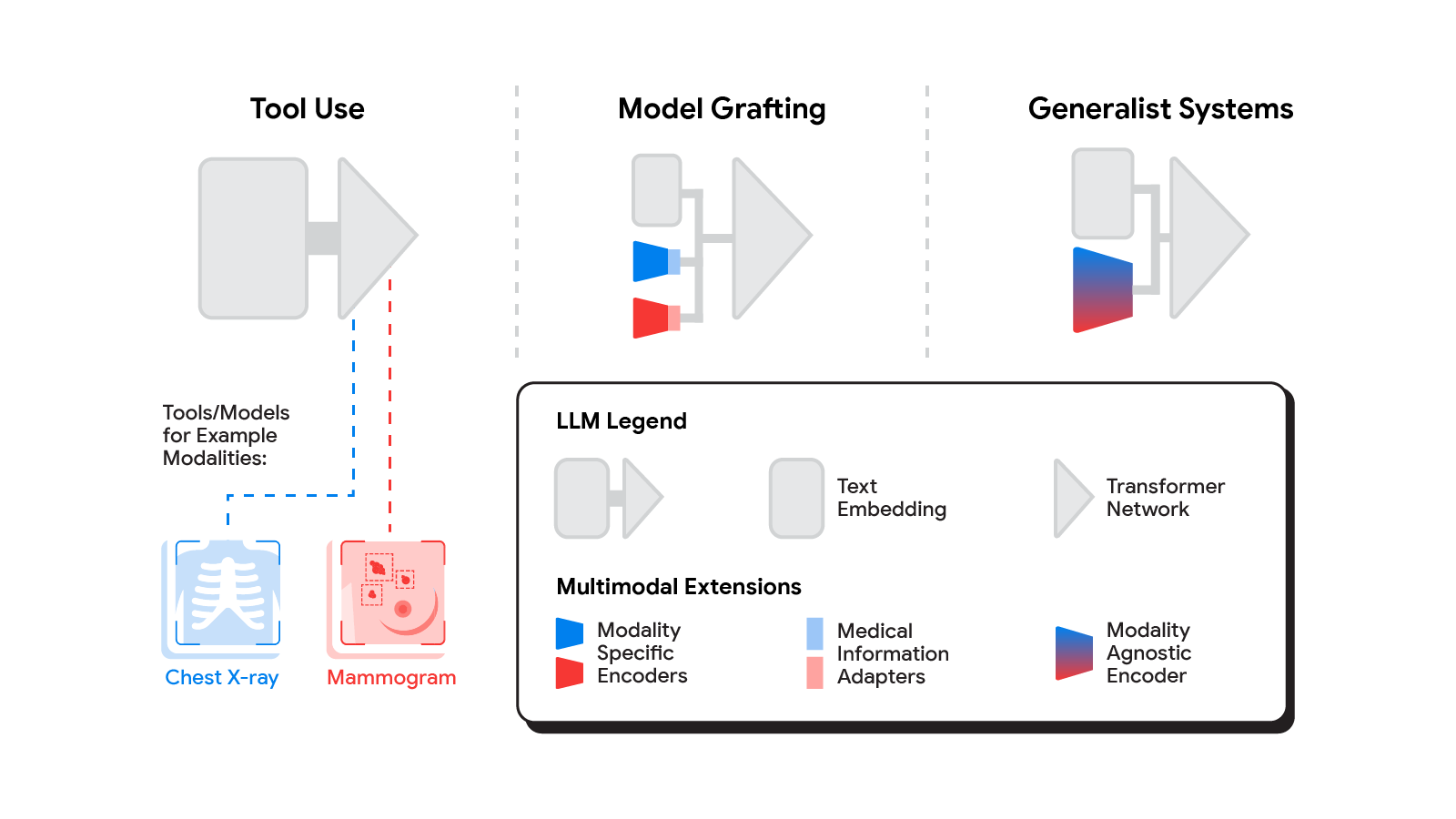
A spectrum of approaches
Several methods for building multimodal LLMs have been proposed in recent months [1, 2, 3], and no doubt new methods will continue to emerge for some time. For the purpose of understanding the opportunities to bring new modalities to medical AI systems, we’ll consider three broadly defined approaches: tool use, model grafting, and generalist systems.
Tool use
In the tool use approach, one central medical LLM outsources analysis of data in various modalities to a set of software subsystems independently optimized for those tasks: the tools. The common mnemonic example of tool use is teaching an LLM to use a calculator rather than do arithmetic on its own. In the medical space, a medical LLM faced with a chest X-ray could forward that image to a radiology AI system and integrate that response. This could be accomplished via application programming interfaces (APIs) offered by subsystems, or more fancifully, two medical AI systems with different specializations engaging in a conversation.
This approach has some important benefits. It allows maximum flexibility and independence between subsystems, enabling health systems to mix and match products between tech providers based on validated performance characteristics of subsystems. Moreover, human-readable communication channels between subsystems maximize auditability and debuggability. That said, getting the communication right between independent subsystems can be tricky, narrowing the information transfer, or exposing a risk of miscommunication and information loss.
Model grafting
A more integrated approach would be to take a neural network specialized for each relevant domain, and adapt it to plug directly into the LLM — grafting the visual model onto the core reasoning agent. In contrast to tool use where the specific tool(s) used are determined by the LLM, in model grafting the researchers may choose to use, refine, or develop specific models during development. In two recent papers from Google Research, we show that this is in fact feasible. Neural LLMs typically process text by first mapping words into a vector embedding space. Both papers build on the idea of mapping data from a new modality into the input word embedding space already familiar to the LLM. The first paper, “Multimodal LLMs for health grounded in individual-specific data”, shows that asthma risk prediction in the UK Biobank can be improved if we first train a neural network classifier to interpret spirograms (a modality used to assess breathing ability) and then adapt the output of that network to serve as input into the LLM.
The second paper, “ELIXR: Towards a general purpose X-ray artificial intelligence system through alignment of large language models and radiology vision encoders”, takes this same tack, but applies it to full-scale image encoder models in radiology. Starting with a foundation model for understanding chest X-rays, already shown to be a good basis for building a variety of classifiers in this modality, this paper describes training a lightweight medical information adapter that re-expresses the top layer output of the foundation model as a series of tokens in the LLM’s input embeddings space. Despite fine-tuning neither the visual encoder nor the language model, the resulting system displays capabilities it wasn’t trained for, including semantic search and visual question answering.
 |
| Our approach to grafting a model works by training a medical information adapter that maps the output of an existing or refined image encoder into an LLM-understandable form. |
Model grafting has a number of advantages. It uses relatively modest computational resources to train the adapter layers but allows the LLM to build on existing highly-optimized and validated models in each data domain. The modularization of the problem into encoder, adapter, and LLM components can also facilitate testing and debugging of individual software components when developing and deploying such a system. The corresponding disadvantages are that the communication between the specialist encoder and the LLM is no longer human readable (being a series of high dimensional vectors), and the grafting procedure requires building a new adapter for not just every domain-specific encoder, but also every revision of each of those encoders.
Generalist systems
The most radical approach to multimodal medical AI is to build one integrated, fully generalist system natively capable of absorbing information from all sources. In our third paper in this area, “Towards Generalist Biomedical AI”, rather than having separate encoders and adapters for each data modality, we build on PaLM-E, a recently published multimodal model that is itself a combination of a single LLM (PaLM) and a single vision encoder (ViT). In this set up, text and tabular data modalities are covered by the LLM text encoder, but now all other data are treated as an image and fed to the vision encoder.
 |
| Med-PaLM M is a large multimodal generative model that flexibly encodes and interprets biomedical data including clinical language, imaging, and genomics with the same model weights. |
We specialize PaLM-E to the medical domain by fine-tuning the complete set of model parameters on medical datasets described in the paper. The resulting generalist medical AI system is a multimodal version of Med-PaLM that we call Med-PaLM M. The flexible multimodal sequence-to-sequence architecture allows us to interleave various types of multimodal biomedical information in a single interaction. To the best of our knowledge, it is the first demonstration of a single unified model that can interpret multimodal biomedical data and handle a diverse range of tasks using the same set of model weights across all tasks (detailed evaluations in the paper).
This generalist-system approach to multimodality is both the most ambitious and simultaneously most elegant of the approaches we describe. In principle, this direct approach maximizes flexibility and information transfer between modalities. With no APIs to maintain compatibility across and no proliferation of adapter layers, the generalist approach has arguably the simplest design. But that same elegance is also the source of some of its disadvantages. Computational costs are often higher, and with a unitary vision encoder serving a wide range of modalities, domain specialization or system debuggability could suffer.
The reality of multimodal medical AI
To make the most of AI in medicine, we’ll need to combine the strength of expert systems trained with predictive AI with the flexibility made possible through generative AI. Which approach (or combination of approaches) will be most useful in the field depends on a multitude of as-yet unassessed factors. Is the flexibility and simplicity of a generalist model more valuable than the modularity of model grafting or tool use? Which approach gives the highest quality results for a specific real-world use case? Is the preferred approach different for supporting medical research or medical education vs. augmenting medical practice? Answering these questions will require ongoing rigorous empirical research and continued direct collaboration with healthcare providers, medical institutions, government entities, and healthcare industry partners broadly. We look forward to finding the answers together.
Article Source link and Credit